Time-series MODIS data analysis using TIMESAT
Timesat software provides means for efficient estimation of vegetation parameters, this article is about how to use it with MODIS time series data.
About TIMESAT
TIMESAT is the program designed to extract seasonal information from any kind of time-series. It allows to obtain information on phenological parameters (e.g. beginning of season, end of season, amplitude, etc.), with outputs in the form of:
- ASCII text file – for single pixels processing or data series;
- raster layers – for spatial dataset input. Working with MODIS data, we will mostly have to deal with this type of data.
The TIMESAT program can be downloaded from (authors consent required): http://www.nateko.lu.se/personal/Lars.Eklundh/TIMESAT/timesat.html
All software and data are stored in pkzip compressed format and need to be unzipped into an empty folder. No changes need to be made to the registry or other system files, no additional installation steps should be performed. The TIMESAT folder (currently timesat_2_3) contains six subdirectories:
data |
Example databases |
documentation |
User manuals and technical papers |
run |
Working folder |
timesat_fortran |
Binaries (MSDOS-Win32) and source code (in Compaq Fortran 90) for time-series seasonal data extraction |
timesat_matlab |
Graphical Interface |
tools |
Binaries (MSDOS-Win32) and source code (in fortran 90) for getting raw output images with the seasonal parameters/actual calculation |
TIMESAT is able to read and write raw or flat binary files, one file per image. Currently 8-bit unsigned integer, 16-bit signed integer or 32-bit signed real data are supported.
TIMESAT software is composed by three groups of programs:
a) Timesat Graphical User Interface (TimesatGUI)
b) MS-DOS TIMESAT software
c) tools for managing and converting output data to proper raw images (BIL). This output is supposed to be processed in any image processing software.
TimesatGUI
TimesatGUI acts as a visual interface, which helps to carry out exploratory analysis of the data, it processes data series for single pixels to select proper parameters which will be further used to generate output matrices.
Current version of TimesatGUI is 2.3, it was developed under Matlab v. 7.1, but also runs under Matlab v. 6.5.
TIMESAT is able to read and write raw binary files in BIL format, one file per image, no layer stacks are necessary as input. Currently 8-bit unsigned integer, 16-bit signed integer or 32-bit signed real data are supported.
More information about TimesatGUI with examples can be found in the manual, which can be found in “timesat2_3/documentation” folder.

MODIS Data
Downloading and preparation
To get started the user needs to get MODIS time-series data from http://edcimswww.cr.usgs.gov/pub/imswelcome/ and convert them to BIL format (more about conversion).
It is also necessary to add in the images folder a text file with the number of files to process in the first row and their path and names in the next rows. TimesatGUI manual suggest using the folder “data” for this purpose. The structure of the text file is:
..\data\wa\wa_nd98011.img
..\data\wa\wa_nd98012.img
..\data\wa\wa_nd98013.img
..\data\wa\wa_nd98021.img
...
..\data\wa\wa_nd98119.img
A specific format for the name or extension is not required, so images can have “.bil” extensions or don't have extension at all. Neither is necessary to have a header, TimesatGUI is asking you information about number of rows and columns every time you are running it independently of existing header.
TIMESAT supports both relative and absolute paths, but using relative paths you should run TIMESAT software from the proper working folder to the “data” folder (using TimesatGUI you need to configure the working folder prior to start the TimesatGUI routine). TimesatGUI manual suggest using the folder “run” for this purpose.
Loading and fitting
Loading and processing large files is troublesome, so it is recommended that you clip your data to a common extent before using it, this can be easily done using clip-all.aml in Arcinfo Workstation or any other software. You can also set an analysis window for your data using TimesatGUI itself.
The function fitting is done in several steps, on each of them, you'll need to specify certain values:
- Number of seasons and their approximate timing is defined.
- Filters for the data applied or fits smooth functions to the data (Savitzky-Golay filter, or least-squares fitted Gaussian or logistic smooth functions).
- After the fitting has been achieved, the seasonality parameters are computed and written to output files.
TIMESAT allows to use ancillary data for assigning weights to the values in the time-series, it is useful if we have a quality assessment data associated with every image in the time-series (each MODIS product has one or several QA band). The quality values for every single pixel are up to the user, but the authors suggest to use values from 1 for clear conditions to 0 for cloudy/contaminated conditions. The quality files are to be read by TimesatGUI in the same way as the time-series image, in BIL format with a separate lsit file which should contain the number of images in the first row and the path and name of the files in the subsequent rows.
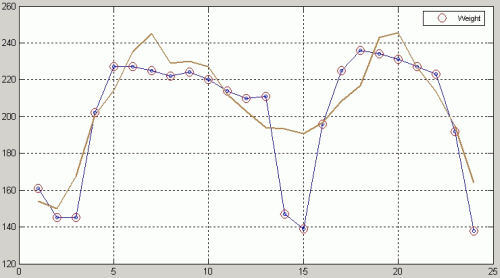
Select Write to parameter.txt option from the menu to write the values to the file parameter.txt in your current working Matlab directory. This file contains all the parameters you've provided on the previous steps:

Generating output phenologies and fits
Additional utilities are used for getting information, fitting parameters and phenologies calculations from your time-series data. These tools are located under the “timesat_fortran” folder. The two binaries to be used are: timesatseries and timesatimage.
The first program reads formatted list of time-series parameters generated on the previous step (parameters.txt) and the second program reads data files itself (BILs). These tools work in exactly the same way as the TimesatGUI, but they need to be run from MS-DOS console. You can also modify parameters directly once you know the meaning of them by changing the values in the “parameter.txt” file using any text editor.
To run these tools, you need to start MS-DOS session: select Start\Run...\ and type in cmd. After it run timesatimage program specifying full path to the executable, for example:
In this example parameter.txt is located in the same directory as timesatimage.exe. It can also be stored in any other folder, in this case you'll need to specify the path it completely, for example:
As an output we'll have seven files divided in 3 groups, 8 files total, N - job name specified in parameters.txt:
- Sensor data and fitted functions (sensordata_N, fitSG_N, fitAG_N, fitDL_N). Three fits files are associated with three possible methods: SG - Savitsky-Golay, AG - Asymmetric Gaussian, DL - Logistic
- Phenological parameters (phenologySG_N, phenologyAG_N, phenologyDL_N). Three files are associated with three possible methods: SG - Savitsky-Golay, AG - Asymmetric Gaussian, DL - Logistic
- Parameter file copy (input_N.txt)
These files contain all the information per pixel for all our time-series, but they are not in raw image (BIL) format yet. To do so we need to use the third group of programs (tools)
Generating output rasters
Using the utility phen2img from tools folder, phenology data created on the previous step can be converted to images. To do this, in DOS session window:
After this, tolls will ask you for the name of the phenology file, created on the previous step, enter it, for example:
You'll be asked to select any of 11 parameters of vegetation which can be calculated:
- Start of season
- End of season
- Length of season
- Base value
- Position of middle of season
- Maximum of fitted data
- Amplitude
- Left derivative
- Right derivative
- Large integral
- Small integral
For any parameters selected, some additional information will be requested by the tool:
- Min and max dates for season to process
- Code for missing season within min and max dates - if no complete season is found within the time span the code for missing season will be written to the corresponding pixel in the image.
- Code for missing phenology data for pixel - if no phenology information is found for the pixel (e.g. over oceans or where there are too many missing values) the code for missing phenology data will be written to the pixel in the image.
- Name of output files, file type for the output image files (signed 16-bit or real 32-bit).
Organization of output files
You would need to run this tool as many times as many seasonal parameters you want to calculate. There can be one or more seasons in your data (this should be same as the number you specified in you parameter.txt). If you have 2 seasons, then two files for each seasonal parameter is being created with _s1 and _s2 ending. And the name of the file equals to the value, you've specified already as a output.
Each seasonal parameter will be represented as a raster, one for each season. In terms of time period used, you can specify the time range for which you want to derive seasonal information. So, for example, you can do it for 3 years of data, or just one, by specifying Image scaling interval option of phen2img.
Total potential number of files that can be generated depends of:
- how many seasons do you have in your data (1-2)
- how many seasonal parameters do you need (1-11)
- which fitting algorithm did you use (1-3)
- what time period you want (each month, each year, average for all years, etc.)
The results are stored in the place as seasonal parameters files and fits are stored.
The resulting image files can be displayed using any imaging processing software capable of handling flat binary files. The output images are given without header and georeferencing, a simple procedure to restore it and be able to display the images is to copy a header and georeferencing file from any of the input images and rename it with the name of the desired output image.
For example: if you have as input these files:
input.bil
input.blw
input.hdr
input.stx
You'll need to copy BLW and HDR file to match the name of the output file generated by timesatimage. After this you will have these files:
output.bil
output.blw
output.hdr
Source MODIS data is 16-bit, but timesatimage outputs can be also 32-bit, if, while generating vegetation parameters file you've chosen 32-bit images as output, it is also necessary to edit the HDR file to reflect the changes in the number of bits, for this:
- Open HDR file with any text editor
- Find the string saying NBITS 16
- Change it to NBITS 32
- Find the strings BANDROWBYTES and TOTALROWBYTES
- The number after this string should twice the initial value. So it for the source data it says:
BANDROWBYTES 8640 for output it should be BANDROWBYTES 17280.
After loading data and setting up a classified legend, your results can look like this:
 |
 |
 |
 |
 |
 |
Seasonal amplitude - difference between the peak value and the base level. |
Peak - largest data value during the season. |
Start of season - time for which the left edge has increased to a user defined level (often 20% of the seasonal amplitude) measured from the left minimum level. |
|||
Обсудить в форуме Комментариев — 5
Последнее обновление: September 09 2021
© GIS-Lab и авторы, 2002-2021. При использовании материалов сайта, ссылка на GIS-Lab и авторов обязательна. Содержание материалов - ответственность авторов. (подробнее).


